SumPubMed: Summarization Dataset of PubMed Scientific Articles
2021
https://doi.org/10.18653/V1/2021.ACL-SRW.30…
12 pages
1 file
Sign up for access to the world's latest research
Abstract
Most earlier work on text summarization is carried out on news article datasets. The summary in these datasets is naturally located at the beginning of the text. Hence, a model can spuriously utilize this correlation for summary generation instead of truly learning to summarize. To address this issue, we constructed a new dataset, SUMPUBMED, using scientific articles from the PubMed archive. We conducted a human analysis of summary coverage, redundancy, readability, coherence, and informativeness on SUMPUBMED. SUMPUBMED is challenging because (a) the summary is distributed throughout the text (not-localized on top), and (b) it contains rare domain-specific scientific terms. We observe that seq2seq models that adequately summarize news articles struggle to summarize SUMPUBMED. Thus, SUMPUBMED opens new avenues for the future improvement of models as well as the development of new evaluation metrics.
FAQs
AI
What are the main characteristics of the SUMPUBMED dataset?add
SUMPUBMED consists of 33,772 biomedical articles, averaging 4,000 words each, from diverse medical literature. It emphasizes non-localized summarization, making it distinct from typical short news articles.
How does SUMPUBMED evaluate summary quality compared to previous datasets?add
The study finds significant differences in summary evaluation, noting that ROUGE scores correlate poorly with human assessments on SUMPUBMED. This indicates a need for new metrics tailored to scientific summarization.
What preprocessing techniques were implemented in the creation of SUMPUBMED?add
The dataset underwent extensive preprocessing, removing non-textual elements like figures and citations, resulting in succinct but informative content. This level of preprocessing is emphasized as a key differentiator from other datasets.
Which summarization models were evaluated on the SUMPUBMED dataset?add
The research assesses multiple models including extractive, abstractive (seq2seq with attention), and hybrid methods. Each method's performance was evaluated based on ROUGE metrics and human quality assessments.
What do the findings suggest about the effectiveness of hybrid summarization approaches?add
Results indicate that hybrid approaches, combining extractive and abstractive techniques, reduce redundancy and improve summary coherence significantly. Specifically, using coverage mechanisms enhanced summary quality in complex biomedical texts.






Related papers
Proceedings of the Second Workshop on Scholarly Document Processing, 2021
Large pretrained models have seen enormous success in extractive summarization tasks. We investigate, here, the influence of pretraining on a BERT-based extractive summarization system for scientific documents. We derive performance improvements using an intermediate pretraining step that leverages existing summarization datasets and report state-of-theart results on a recently released scientific summarization dataset, SCITLDR. We systematically analyze the intermediate pretraining step by varying the size and domain of the pretraining corpus, changing the length of the input sequence in the target task and varying target tasks. We also investigate how intermediate pretraining interacts with contextualized word embeddings trained on different domains.
2020 IEEE International Conference on Big Data (Big Data), 2020
Recent advances in natural language processing have enabled automation of a wide range of tasks, including machine translation, named entity recognition, and sentiment analysis. Automated summarization of documents, or groups of documents, however, has remained elusive, with many efforts limited to extraction of keywords, key phrases, or key sentences. Accurate abstractive summarization has yet to be achieved due to the inherent difficulty of the problem, and limited availability of training data. In this paper, we propose a topic-centric unsupervised multi-document summarization framework to generate extractive and abstractive summaries for groups of scientific articles across 20 Fields of Study (FoS) in Microsoft Academic Graph (MAG) and news articles from DUC-2004 Task 2. The proposed algorithm generates an abstractive summary by developing salient language unit selection and text generation techniques. Our approach matches the state-of-the-art when evaluated on automated extract...
Bulletin of Electrical Engineering and Informatics, 2022
Along with the increasing number of scientific publications, many scientific communities must read the entire text to get the essence of information from a journal article. This will be quite inconvenient if the scientific journal article is quite long and there are more than one journals. Motivated by this problem, encourages the need for a method of text summarization that can automatically, concisely, and accurately summarize a scientific article document. The purpose of this research is to create an extractive text summarization by doing feature engineering to extract the semantic information from the original text. Comparing the long short-term memory algorithm and gated recurrent units and were used to get the most relevant sentences to be served as a summary. The results showed that both algorithms yielded relatively similar accuracy results, with gated recurrent units at 98.40% and long short-term memory at 98.68%. The evaluation method with matrix recall-oriented understudy for gisting evaluation (ROUGE) is used to evaluate the summary results. The summary results produced by the LSTM model compared to the summary results using the latent semantic analysis (LSA) method were then obtained recall values at ROUGE-1, ROUGE-2, and ROUGE-L respectively were 76.25%, 59.49%, and 72.72%.
2016
Readers usually rely on abstracts to identify relevant medical information from scientific articles. Abstracts are also essential to advanced information retrieval methods. More than 50 thousand scientific publications in PubMed lack author-generated abstracts, and the relevancy judgements for these papers have to be based on their titles alone. In this paper, we propose a hybrid summarization technique that aims to select the most pertinent sentences from articles to generate an extractive summary in lieu of a missing abstract. We combine i) health outcome detection, ii) keyphrase extraction, and iii) textual entailment recognition between sentences. We evaluate our hybrid approach and analyze the improvements of multi-factor summarization over techniques that rely on a single method, using a collection of 295 manually generated reference summaries. The obtained results show that the hybrid approach outperforms the baseline techniques with an improvement of 13% in recall and 4% in ...
The 22nd Workshop on Biomedical Natural Language Processing and BioNLP Shared Tasks
In this paper, we consider the challenge of summarizing patients' medical progress notes in a limited data setting. For the Problem List Summarization (shared task 1A) at the BioNLP Workshop 2023, we demonstrate that Clinical-T5 fine-tuned to 765 medical clinic notes outperforms other extractive, abstractive and zeroshot baselines, yielding reasonable baseline systems for medical note summarization. Further, we introduce Hierarchical Ensemble of Summarization Models (HESM), consisting of tokenlevel ensembles of diverse fine-tuned Clinical-T5 models, followed by Minimum Bayes Risk (MBR) decoding. Our HESM approach lead to a considerable summarization performance boost, and when evaluated on held-out challenge data achieved a ROUGE-L of 32.77, which was the best-performing system at the top of the shared task leaderboard. 1
arXiv (Cornell University), 2024
This work presents a dynamic vocabulary adaptation strategy, MEDVOC, for fine-tuning pretrained language models (PLMs) like BertSum-Abs, BART, and PEGASUS for improved medical text summarization. In contrast to existing domain adaptation approaches in summarization, MEDVOC treats vocabulary as an optimizable parameter and optimizes the PLM vocabulary based on fragment score conditioned only on the downstream task's reference summaries. Unlike previous works on vocabulary adaptation (limited only to classification tasks), optimizing vocabulary based on summarization tasks requires an extremely costly intermediate fine-tuning step on large summarization datasets. To that end, our novel fragment score-based hyperparameter search very significantly reduces this fine-tuning timefrom 450 days to less than 2 days on average. Furthermore, while previous works on vocabulary adaptation are often primarily tied to single PLMs, MEDVOC is designed to be deployable across multiple PLMs (with varying model vocabulary sizes, pre-training objectives, and model sizes)-bridging the limited vocabulary overlap between the biomedical literature domain and PLMs. MEDVOC outperforms baselines by 15.74% in terms of Rouge-L in zero-shot setting and shows gains of 17.29% in high Out-Of-Vocabulary (OOV) concentrations. Our human evaluation shows MEDVOC generates more faithful medical summaries (88% compared to 59% in baselines).
Lecture Notes in Computer Science, 2013
This paper describes experiments to adapt document summarization to the medical domain. Our summarizer combines linguistic features corresponding to text fragments (typically sentences) and applies a machine learning approach to extract the most important text fragments from a document to form a summary. The generic features comprise features used in previous research on summarization. We propose to adapt the summarizer to the medical domain by adding domainspecific features. We explore two types of additional features: medical domain features and semantic features. The evaluation of the summarizer is based on medical articles and targets different aspects: i) the classification of text fragments into ones which are important and ones which are unimportant for a summary; ii) analyzing the effect of each feature on the performance; and iii) system improvement over our baseline summarizer when adding features for domain adaptation. Evaluation metrics include accuracy for training the sentence extraction and the ROUGE measure computed for reference summaries. We achieve an accuracy of 84.16% on medical balanced training data by using an IB1 classifier. Training on unbalanced data achieves higher accuracy than training on balanced data. Domain adaptation using all domain-specific features outperforms the baseline summarization wrt. ROUGE scores, which shows the successful domain adaptation with simple means.
Abstract We perform a quantitative analysis of data in a corpus that specialises on summarisation for Evidence Based Medicine (EBM). The intent of the analysis is to discover possible directions for performing automatic evidence-based summarisation. Our analysis attempts to ascertain the extent to which good, evidence-based, multidocument summaries can be obtained from individual single-document summaries of the source texts.
2016
This work is (partly) supported by the Spanish Ministry of Economy and Competitiveness under the Maria de Maeztu Units of Excellence Programme (MDM-2015-0502), the TUNER project (TIN2015-65308-C5-5-R, MINECO/FEDER, UE) and the European Project Dr. Inventor (FP7-ICT-2013.8.1 - Grant no: 611383).
SUMPUBMED: Summarization Dataset of PubMed Scientific Articles
Vivek Gupta
University of Utah
vgupta@cs.utah.edu
Pegah Nokhiz
University of Utah
pnokhiz@cs.utah.edu
Prerna Bharti
Microsoft Corporation
prerna.bharti@microsoft.com
Abstract
Most earlier work on text summarization is carried out on news article datasets. The summary in these datasets is naturally located at the beginning of the text. Hence, a model can spuriously utilize this correlation for summary generation instead of truly learning to summarize. To address this issue, we constructed a new dataset, SUMPubMED, using scientific articles from the PubMed archive. We conducted a human analysis of summary coverage, redundancy, readability, coherence, and informativeness on SUMPubMED. SUMPubMED is challenging because (a) the summary is distributed throughout the text (not-localized on top), and (b) it contains rare domain-specific scientific terms. We observe that seq2seq models that adequately summarize news articles struggle to summarize SUMPUBMED. Thus, SUMPUBMED opens new avenues for the future improvement of models as well as the development of new evaluation metrics.
1 Introduction
Most of the existing summarization datasets, i.e., CNN Daily Mail and DUC are news article datasets. That is, the article acts as a document, and the summary is a short (10-15 lines) manually written highlight (i.e., headlines). In many cases, these highlights have significant lexical overlap with the few lines at the top of the article. Thus, any model which can extract the top few lines, e.g., extractive methods, performs adequately on these datasets.
However, the task of summarization is not merely limited to short-length news articles. One could also summarize long and complex documents such as essays, research papers, and books. In such cases, an extractive approach will most likely fail. For successful summarization on these documents, one needs to (a) find information from the distributed (non-localized) locale in the large
Harish Karnick
IIT Kanpur
hkarnick@cs.iitk.ac.in
text, (b) perform paraphrasing, simplifying, and shortening of longer sentences and © combine information from multiple sentences to generate the summary. Hence, an abstractive approach will perform better on such large documents.
One obvious source that contains such complex documents is the MEDLINE biomedical scientific articles, which are publicly available. Furthermore, these articles are accompanied by abstracts and conclusions which summarize the documents. Therefore, we constructed a scientific summarization dataset from pre-processed PubMed articles, named SUMPubMED. In comparison to the previous news-article based datasets, SUMPUBMED documents are longer, and the corresponding summaries cannot be extracted by selecting a few sentences from fixed locations in the document.
The dataset, along with associated scripts, are available at https://github.com/vgupta123/ sumpubmed. Our contributions in this paper are:
- We created a new scientific summarization dataset, SUMPUBMED, which has longer text documents and summaries with non-localized information from documents.
- We analyzed the quality of summaries in SUMPubMED on the basis of four parameters: readability, coherence, non-repetition, and informativeness using human evaluation.
- We evaluated several extractive, abstractive (seq2seq), and hybrid summarization models on SUMPubMED. The results show that SUMPubMED is more challenging compared to the earlier news-based datasets.
- Lastly, we showed that the standard summarization evaluation metric, ROUGE (Lin, 2004), correlates poorly with human evaluations on SUMPUBMED. This indicates the
need for a new evaluation metric for the scientific summarization task.
In Section 1, we provided a brief introduction. The remaining parts of the paper are organized as follows: in Section 2 we explain how SumPubMed was created. In Section 3, we explain how summaries were annotated by human experts. We then move on to experiments in Section 4. We next discuss the results and analysis in Section 5, followed by the related work in Section 6. Lastly, we move on to the conclusions in final Section 7.
2 SumPubMed Creation
SUMPubMED is created from PubMed biomedical research papers, which has 26 million documents. The documents are sourced from diverse literature, including MEDLINE, life science journals, and online books. For SumPubMed creation we took 33,772 documents from Bio Med Central (BMC). BMC incorporates research papers related to medicine, pharmacy, nursing, dentistry, health care, health services, etc.
The research documents in BMC contain two subsections: Front and Body. The front part of the document is basically the abstract and taken as the gold summary. The body part which is taken as the main document contains three subsections: background, results, and conclusion.
Preprocessing The average word count in the PubMed scientific articles is around 4,000 words for each document and 250 to 300 lines in every document. Therefore, to create SumPubMed, we performed extensive preprocessing so that nontextual content is removed and the overall text is reduced to a more manageable size. This extensive pre-processing step is one of the main factors that sets SUMPubMed apart from similar datasets (Cohan et al., 2018).
During preprocessing, the non-textual content from the text was removed by: (a) replacing citations and digits in the content with < cit > and < dig > labels, (b) removing figures, tables, signatures, subscripts, superscripts, and their associated text (e.g., captions), and © removing the acknowledgments and references from the text. All the preprocessing was done on a sentence level utilizing the Python regex library. 1 After preprocessing,
[1]we convert the final document to an XML format and use the SAX parser to parse it.
SAX vs DOM parser: In SAX, events are triggered when the XML is being parsed. When the parser is parsing the XML and encounters a tag starting (e.g., < something > ), then it triggers the tagStarted event (actual name of the event might differ). Similarly, when the end of the tag is met while parsing ( < /something > ), it triggers tagEnded. Using a SAX parser implies one needs to handle these events and make sense of the data returned with each event. One could also use the DOM parser, 2 where no events are triggered while parsing. In DOM the entire XML is parsed, and a DOM tree (of the nodes in the XML) is generated and returned. In general, DOM is easier to use but has a huge overhead of parsing the entire XML before one can start using it; therefore, we use SAX instead.
An example of the front part, body part, and the XML file formed from the pre-processed text is shown in https://github.com/vguptal23/ sumpubmed/blob/master/template.pdf.
Versions of SUMPubMed We maintained three versions of SUMPubMed with varying degrees of preprocessing, a) XML, b) Raw Text, and c) Nounphrases. Details of each version are as follows:
- In the XML version, we exported the whole dataset into a single XML file
- The Raw Text version is obtained after preprocessing when removing non-textual context is completed, followed by XML parsing.
- In the Noun phrases version, we processed the raw text version further to ensure that the summary and the text have the same named entities.
We found that standard Name Entity Recognition (NER) (Finkel et al., 2005) and Biomedical Named Entity Recognizer (ABNER) (Settles, 2005) fail to pick the scientific named entities correctly. Note that the main reason behind ABNER insufficiency is the presence of novel PubMed named entities that were not covered by any of the classes in the ABNER tool. Therefore, we use a simple heuristic of noun intersection between summary and main-text noun phrases to obtain plausible entity sets. This produced a shorter version of both the text and the summary than the original pair.

Figure 1: SUMPUBMED creation pipeline.
| Version | Avg. Stats | Summary | Article |
|---|---|---|---|
| Raw Text | Words | 277 | 4227 |
| version | Sents | 14 | 203 |
| Noun Phrase | Words | 223 | 1578 |
| version | Sents | 10 | 57 |
| Hybrid | Words | 223 | 1891 |
| version | Sents | 10 | 71 |
Table 1: Average number of sentences and words in the abstract and text in the three SUMPUBMED versions
The SUMPUBMED versions statistics is given in Table 1. The SUMPubMED overall creation pipeline is shown in Figure 1.
3 Human Annotation of SUMPubMED
Inspired from work on human evaluation of summaries by Friedrich et al. (2014), we distributed 50 randomly chosen summaries from the noun-phrase versions of SUMPUBMED to 10 expert annotators (graduate NLP students) such that we have 3 annotation for each summary. We asked these humanannotators to rate the summaries on a scale of 1 to 10. We created different document files, each having 10 pairs of summaries where we randomly shuffled between reference and generated summaries with respect to the placement on the page (left or right). The annotators evaluated the summaries based on the following criteria:
- Non-Repetition and no factual Redundancy
(Non-Re): There should not be redundancy in the factual information, and no repetition of sentences is allowed.
- Coherence (Coh): Coherence means “continuity of sense”. The arguments have to be connected sensibly so that the reader can see consecutive sentences as being about one (or a related) concept.
- Readability (Read): Consideration of general readability criteria such as good spelling, correct grammar, understandability, etc. in the summaries.
- Informativeness, Overlap and Focus (IOF): How much information is covered by the summary. The goal is to find the common pieces of information via matching the same keywords (or key phrases), such as “Nematodes”, across the summary. For overlaps, annotators compare the keywords’ (or key-phrases) occurrence frequency and ensure the summaries are on the same topic.
The average scores and standard deviations are shown in Table 2. Annotators found that for readability, coherence, and non-repetitiveness, the quality of summaries is satisfactory. However, for informativeness and overlap, it is hard to evaluate summaries due to domain-specific technical terms.
| Criteria | Mean (μ) | S.D. (σ) |
|---|---|---|
| Non-Re | 7.19 | 0.755 |
| Coh | 6.87 | 0.705 |
| Read | 6.82 | 0.821 |
| IOF | 6.31 | 0.879 |
Table 2: Mean and Standard Deviation (SD) scores of human annotation on 50 summaries
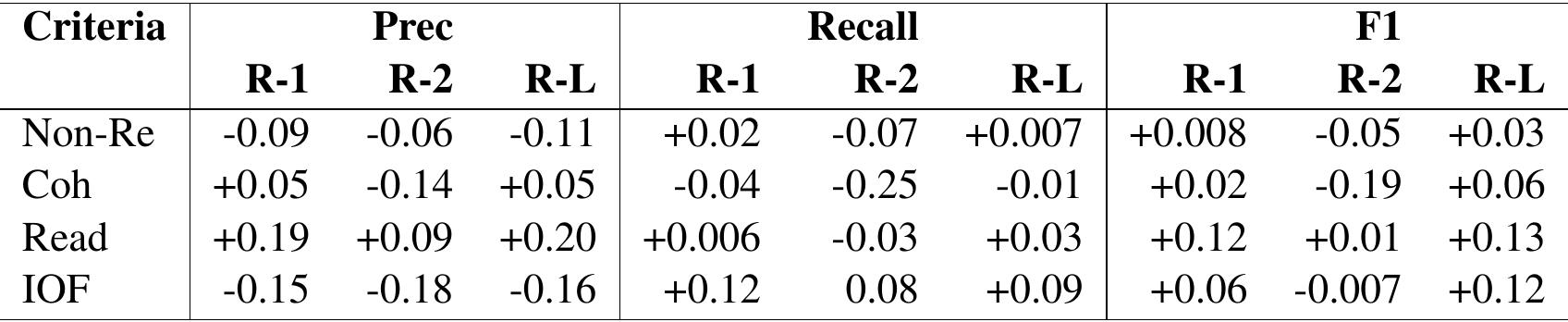
ROUGE and Human Scores For the 50 summaries evaluated by expert annotators, we calculated the Pearson’s correlation (Pearson, 1895) between ROUGE (Lin, 2004) scores (ROUGE-1 (R1), ROUGE-2 (R-2) and ROUGE-L (R-L)) in terms of precision, recall and F1 score with the humanevaluated scores. ROUGE- n is an n-gram similarity measure that computes uni/bi/trigram and higher n-gram overlaps. In R-L, L refers to the Longest Common Subsequence (LCS) overlap: a subsequence of matching words with the maximal length that is common in both texts with the order of words being preserved. Pearson’s correlation value (between -1 and +1 ) quantifies the degree to which quantitative and continuous variables are related to each other. The Pearson’s correlations values are shown in Table 3.
ROUGE scores assume that a high-quality summary generated by a model should have common words and phrases with a gold-standard summary. However, this is not always true because (a) there can be semantically similar meaning (synonymous) word usage, and (b) there can be the usage of text paraphrases (similar information conveyed) with a little lexical overlap in the reference summary text. Therefore, merely considering lexical overlaps to evaluate summary quality is not sufficient. A high ROUGE score may indicate a good summary, but a low ROUGE score does not necessarily indicate a bad summary. Furthermore, while summarizing large documents, humans tend to utilize different paraphrasing/words to convey the same meaning in a shorter form. Several studies by Cohan and Goharian (2016); Dohare et al. (2017) argue that ROUGE is not an accurate estimator of the quality of a summary for scientific input, e.g., biomedical text. Hence, a weak correlation of ROUGE scores with human ratings on SUMPubMed, as reported in Table 3, should not be a surprise. That is, all correlation values in Table 3 are close to zero, so we can conclude that Rouge scores are weakly related with human ratings on the SUMPubMed.
4 Experiments
We have used the noun phrase version of SumPubMed in the abstractive summarization settings and the Hybrid version of SumPubMed in the extractive and the hybrid settings, i.e., (extractive + abstractive) summarizations. We split the dataset into train ( 93% ), test ( 3% ), and validation (4%) sets. Before training, we wrote a script that first tokenizes all input files and then forms the vocabulary and chunked files for the train, test, and validation sets. This step converts the input into a suitable format for the seq2seq models.
4.1 Baseline Models
We use the following models on SUMPubMed for evaluation: We use extractive, abstractive, and hybrid (extractive + abstractive) automatic summarization methods to evaluate SUMPubMed.
Abstractive Methods We use several modifications of seq2seq with attention, as described below:
Seq2Seq with Attention (Nallapati et al., 2016): The encoder is a single layer bidirectional LSTM, while the decoder is a single layer unidirectional LSTM. Both the encoder and decoder have same sized hidden states, with an attention mechanism over the source hidden states and a soft-max layer over the vocabulary to generate the words. We use the same vocabulary for both the encoding and the decoding phase.
Seq2Seq with Pointer Generation Networks (See et al., 2017): The previous model has a computational decoder complexity because each time we have to apply the softmax over the entire vocabulary. The model also outputs an excessive number of UNK tokens (UNK is a special token utilized for out-of-vocabulary words) in the target summary. To address this issue, we use a pointer-generator network (See et al. (2017)) which integrates the basic seq2seq model (with attention) with a copying mechanism (Gu et al. (2016)). We call this model seq2seq for the rest of the paper.
The seq2Seq model with Pointer Generation Networks and Coverage Mechanism (+cov) (Mi et al., 2016): The summaries generated by the model discussed before may show repetition, like generating the same arrangement of words multiple times (e.g., “this bioinformatic approach this bioinformatic approach…” ). This repetition of phrases is prominent when generating multi-line summaries. The solu-
| Criteria | PreC | Recall | F1 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| R-1 | R-2 | R-L | R-1 | R-2 | R-L | R-1 | R-2 | R-L | |
| Non-Re | −0.09 | −0.06 | −0.11 | +0.02 | −0.07 | +0.007 | +0.008 | −0.05 | +0.03 |
| Coh | +0.05 | −0.14 | +0.05 | −0.04 | −0.25 | −0.01 | +0.02 | −0.19 | +0.06 |
| Read | +0.19 | +0.09 | +0.20 | +0.006 | −0.03 | +0.03 | +0.12 | +0.01 | +0.13 |
| IOF | −0.15 | −0.18 | −0.16 | +0.12 | 0.08 | +0.09 | +0.06 | −0.007 | +0.12 |
Table 3: Pearson’s correlation between ROUGE scores and human ratings on SUMPUBMED’s noun-phrase version
tion to the problem of redundancy in summaries in seq2seq models is the coverage mechanism of Mi et al. (2016). This model penalizes repeated word generations by keeping track of the hitherto covered parts using attention distribution.
Extractive Methods There are several existing approaches to extractive summarization, mostly derived from LexRank (Erkan and Radev, 2004), and TextRank (Mihalcea and Tarau, 2004). We use TextRank, which is an unsupervised approach for sentence extraction, and has been used successfully in many NLP applications (Hulth, 2003).
Hybrid Methods (Extractive + Abstractive) We also experimented with the hybrid approach for summarization. First, we used extractive summarization using the TextRank ranking algorithm. We then applied abstractive summarization on the extracted text. We used the pointer-generator networks, followed by the coverage mechanism for the abstractive summarization. In this setting, we have not perfomed any preprocessing before extractive summarization to decrease the length of the documents. The extractive summarization step makes the text length sufficient to apply the abstractive summarization step on it quite easily.
4.2 Experimental Settings
While decoding seq2seq models (for abstractive and hybrid models), we use a beam search (Medress et al., 1977) with a beam width of 4.Note that, Beam search is a greedy technique which chooses the most likely token from all generated tokens at each step to obtain the best b sequences (the hyper-parameter b here represents the beam width). Beam search is shown to be better than generating the first sequence.
We also experimented with varying target summary lengths (i.e., the number of decoding steps) for seq2seq models. We report both seq2seq models with and without coverage results for comparison. We considered ROUGE-1 (R-1), ROUGE-2 (R-2), and ROUGE- L (R-L)'s precision, recall, and
F1 score for evaluation.
Hyper-parameters The hyper-parameters used for the seq2seq model is in Table 4.
| Hyper-parameter | Value |
|---|---|
| LSTM Hidden state size | 256 |
| Word embedding dimensions | 128 |
| Batch Size | 16 |
| encoder steps training | 100−1000 |
| encoder steps testing | 100−4000 |
| decoder steps length | 100−250 |
| beam size | 4 |
| learning rate for adagrad | 0.15 |
| maximum gradient norm | 2.0 |
Table 4: Hyper-parameters for seq2seq models
We utilized tensorflow package 3 for models and ROUGE evaluation package pyrouge 4 for the evaluation metric. We use a single GeForce GTX TITANX with 12 GB GPU memory taking on average 5 to 6 days per model for model training.
5 Results and Analysis
Results on SUMPUBMED for abstractive methods, i.e., seq2seq models (with and without coverage), the extractive method of TextRank, and the hybrid approach, i.e., TextRank + seq2seq (with and without coverage) are shown in Tables 6, 7, and 8, respectively. We also evaluated the seq2seq models on news datasets (CNN/Daily Mail and DUC 2001) for comparison, as shown in Table 5.
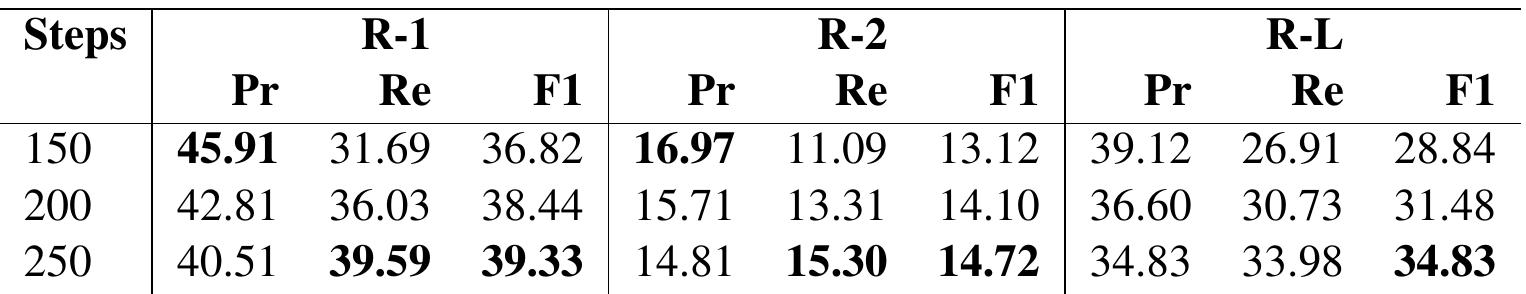
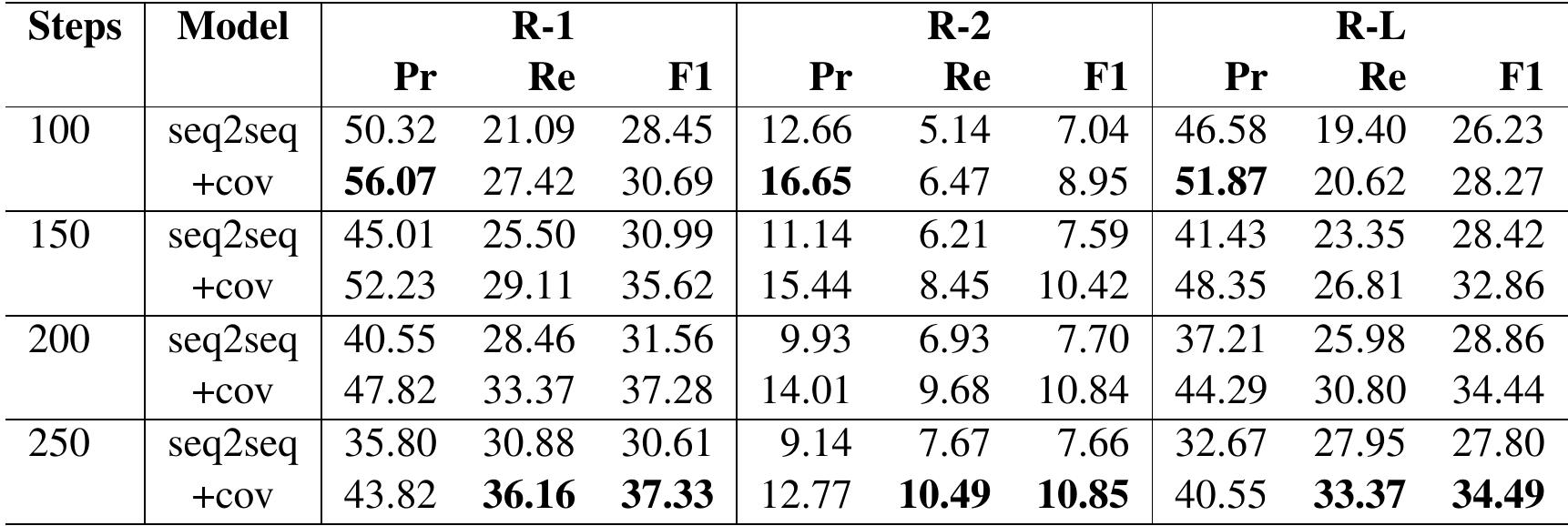
Analysis: In all three approaches, abstractive in Table 6, extractive in Table 7 and hybrid in Table 8 , we notice that the ROUGE Recall and F1-score increase, whereas precision decreases with the number of words ( 100 to 250 ) in the target summaries. The increase in Recall is expected as the chances of lexical overlap are more with larger generated summaries. Precision decreases because, with more
3 https://www.tensorflow.org/
4 https://pypi.org/project/pyrouge/ ↩︎
| Data | Model | R-1 | R-2 | R-L | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pr | Re | F1 | Pr | Re | F1 | Pr | Re | F1 | ||
| CNN | seq2seq | 33.49 | 38.49 | 34.61 | 13.89 | 15.87 | 14.29 | 30.15 | 34.64 | 31.15 |
| -DM | +cov | 38.59 | 41.10 | 38.53 | 16.84 | 17.83 | 16.75 | 35.56 | 37.81 | 35.48 |
| DUC | seq2seq | 41.34 | 21.33 | 27.63 | 14.28 | 7.30 | 9.49 | 32.95 | 16.93 | 21.93 |
| +cov | 43.86 | 21.92 | 28.57 | 15.04 | 7.41 | 9.68 | 34.96 | 17.29 | 22.60 |
Table 5: ROUGE scores on CNN-Dailymail (CNN-DM) and DUC 2001 dataset (DUC) using seq2seq models
| Steps | Model | R-1 | R-2 | R-L | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pr | Re | F1 | Pr | Re | F1 | Pr | Re | F1 | ||
| 100 | seq2seq | 52.30 | 20.56 | 28.01 | 16.01 | 6.17 | 8.50 | 47.97 | 18.70 | 25.53 |
| +cov | 57.50 | 22.66 | 31.04 | 20.28 | 7.74 | 10.73 | 52.62 | 20.56 | 28.23 | |
| 150 | seq2seq | 48.88 | 27.10 | 32.81 | 15.18 | 8.35 | 10.18 | 44.64 | 24.56 | 29.81 |
| +cov | 55.11 | 29.71 | 36.79 | 19.17 | 10.14 | 12.66 | 50.48 | 27.07 | 33.57 | |
| 200 | seq2seq | 44.83 | 30.23 | 33.79 | 13.73 | 9.20 | 10.33 | 40.86 | 27.37 | 30.65 |
| +cov | 52.86 | 33.84 | 39.21 | 18.25 | 11.52 | 13.43 | 48.47 | 30.88 | 35.84 | |
| 250 | seq2seq | 41.18 | 31.84 | 33.00 | 12.80 | 9.79 | 10.22 | 37.68 | 28.89 | 30.03 |
| +cov | 51.11 | 36.24 | 40.13 | 17.63 | 12.39 | 13.77 | 46.92 | 33.13 | 36.73 |
Table 6: ROUGE scores of noun-phrase SUMPUBMED version using a seq2seq model of varying decoding steps
| Steps | R-1 | R-2 | R-L | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pr | Re | F1 | Pr | Re | F1 | Pr | Re | F1 | ||
| 150 | 45.91 | 31.69 | 36.82 | 16.97 | 11.09 | 13.12 | 39.12 | 26.91 | 28.84 | |
| 200 | 42.81 | 36.03 | 38.44 | 15.71 | 13.31 | 14.10 | 36.60 | 30.73 | 31.48 | |
| 250 | 40.51 | 39.59 | 39.33 | 14.81 | 15.30 | 14.72 | 34.83 | 33.98 | 34.83 |
Table 7: Results for TextRank an Extractive Summarization approach on hybrid version of the SUMPUBMED.
| Steps | Model | R-1 | R-2 | R-L | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pr | Re | F1 | Pr | Re | F1 | Pr | Re | F1 | ||
| 100 | seq2seq | 50.32 | 21.09 | 28.45 | 12.66 | 5.14 | 7.04 | 46.58 | 19.40 | 26.23 |
| +cov | 56.07 | 27.42 | 30.69 | 16.65 | 6.47 | 8.95 | 51.87 | 20.62 | 28.27 | |
| 150 | seq2seq | 45.01 | 25.50 | 30.99 | 11.14 | 6.21 | 7.59 | 41.43 | 23.35 | 28.42 |
| +cov | 52.23 | 29.11 | 35.62 | 15.44 | 8.45 | 10.42 | 48.35 | 26.81 | 32.86 | |
| 200 | seq2seq | 40.55 | 28.46 | 31.56 | 9.93 | 6.93 | 7.70 | 37.21 | 25.98 | 28.86 |
| +cov | 47.82 | 33.37 | 37.28 | 14.01 | 9.68 | 10.84 | 44.29 | 30.80 | 34.44 | |
| 250 | seq2seq | 35.80 | 30.88 | 30.61 | 9.14 | 7.67 | 7.66 | 32.67 | 27.95 | 27.80 |
| +cov | 43.82 | 36.16 | 37.33 | 12.77 | 10.49 | 10.85 | 40.55 | 33.37 | 34.49 |
Table 8: ROUGE scores on hybrid version of the SUMPUBMED using Hybrid model: TextRank + seq2seq models
| Model | R-1 | R-2 | R-L | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Pr | Re | F1 | Pr | Re | F1 | Pr | Re | F1 | |
| Abstractive | 51.11 | 36.24 | 40.13 | 17.63 | 12.39 | 13.77 | 46.92 | 33.13 | 36.73 |
| Extractive | 40.51 | 39.59 | 39.33 | 14.81 | 15.30 | 14.72 | 34.83 | 33.98 | 32.82 |
| Hybrid Model | 43.82 | 36.16 | 37.33 | 12.77 | 10.49 | 10.85 | 40.55 | 33.37 | 34.49 |
Table 9: ROUGE comparison on SUMPUBMED. seq2seq abstractive methods’ target summary is of 250 words words, the chances of non-covered words in the output summary also increase.
We notice in both Tables 6 and 8 that by adding
the coverage (+cov) mechanism, the problem of repetition in summaries is solved to a great extent. The ROUGE scores also show improvement after
applying coverage to pointer-generator networks. Thus, one can conclude that pointer generator networks effectively handle named entities and out-of-vocabulary words, and the coverage mechanism is useful to avoid repetitive generation, which is essential for scientific summarization.
In Table 9, we note that in terms of Precision (Pr), the abstractive approach shows the best results. However, the Recall (Re) of the extractive summarization model is always better than abstractive and hybrid approaches. Furthermore, the R-1 Re (ROUGE-1 Recall) and R-L Re (ROUGE-L Recall) for the hybrid models are approximately similar to the abstractive models. We also provide a few qualitative example of summarization on CNN/DailyMail in Appendix Section A, on SumPubMed in Appendic Section B.
6 Related Work
Below, we provide the details of other summarization datasets:
News: CNN-Daily Mail has 92,000 examples with documents of 30 -sentence length with 4 corresponding human-written summaries of 50 words. DUC (Document Understanding Conference), another dataset, contains 500 documents ( 35.6 tokens on average) and summaries ( 10.4 tokens). Gigaword (Rush et al., 2015) has 31.4 document tokens and 8.3 summary tokens. Lastly, X-Sum (Extreme Summarization) (Narayan et al., 2018) contains 20 -sentence ( BBC articles) ( 431 words) and corresponding one-sentence ( 23 words) summaries.
Social Media: Webis-TLDR-17 Corpus (Völske et al., 2017) is a large-scale dataset of 3 million pairs of content and self-written summaries obtained from social media (Reddit). Webis-Snippet20 Corpus (Chen et al., 2020) contains 10 million (webpage content and abstractive snippet) pairs and 3.5 million triples (query terms, abstractive snippets, etc.) for query-based abstractive snippet generation of web pages.
Scientific: Recently, Sharma et al. (2019) released a large dataset of 1.3 million of U.S. patent documents along with human written summaries. However, the closest datasets to SumPubMed are released by Cohan et al. (2018); Kedzie et al. (2018); Gidiotis and Tsoumakas (2019).
Comparison with SumPubMed: News datasets’ summary is located at the top of
the article for most examples. Social media datasets lack the scientific aspect, i.e., complex domain-specific vocabulary and non-localized distributed information of SumpubMed. Other works on the scientific datasets are by Cohan et al. (2018); Kedzie et al. (2018); Gidiotis and Tsoumakas (2019). The closest work to our approach is the PubMed dataset by Cohan et al. (2018). However, unlike SumPubMed, (a) no extensive preprocessing pipeline was applied to clean the text (b) a single version is released compared with SumPubMed’s several versions with distinct properties (varying summary lengths, article lengths, and vocabulary sizes), © only level-1 section headings instead of the whole PubMed document are used, and (d) there is a lack of human evaluation to assess data quality. However, Cohan et al. (2018) do act as an powerful inspiration for our work.
7 Conclusion
We created a non-news, SumPubMed dataset, from the PubMed archive to study how various summarization techniques perform on task of scientific summarization on domain specific scientific texts. These texts have essential information scattered throughout the whole text. In contrast, earlier datasets with news stories appear to mostly have useful information in the first few lines of the document text. We also conducted a human evaluation on aspects such as repetition, readability, coherence, and Informativeness for 50 summaries of 250 words. Each summary is evaluated by 3 different individuals on the basis of four parameters: readability, coherence, non-repetition, and informativeness. Due to the unavailability of any state-of-the-art results on this new dataset, we built several baseline models (extractive, abstractive, and hybrid model) for SumPubMed. To check the significance of our results, we studied the effectiveness of ROUGE through Pearson’s correlation analysis with human-evaluation and observed that many variants of ROUGE scores correlate poorly with human evaluation. Our results indicate that ROUGE is possibly not a proper metric for SumPubMed.
Acknowledgements
We would like to thank the ACL SRW anonymous reviewers for their useful feedback, comments, and suggestions.
References
Wei-Fan Chen, Shahbaz Syed, Benno Stein, Matthias Hagen, and Martin Potthast. 2020. Abstractive snippet generation. In Proceedings of The Web Conference 2020, pages 1309-1319.
Arman Cohan, Franck Dernoncourt, Doo Soon Kim, Trung Bui, Seokhwan Kim, Walter Chang, and Nazli Goharian. 2018. A discourse-aware attention model for abstractive summarization of long documents. In Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 2 (Short Papers), volume 2, pages 615-621.
Arman Cohan and Nazli Goharian. 2016. Revisiting summarization evaluation for scientific articles. In Proceedings of the Tenth International Conference on Language Resources and Evaluation (LREC’16), pages 806-813.
Shibhansh Dohare, Harish Karnick, and Vivek Gupta. 2017. Text summarization using abstract meaning representation. arXiv preprint arXiv:1706.01678.
Günes Erkan and Dragomir R Radev. 2004. Lexrank: Graph-based lexical centrality as salience in text summarization. Journal of artificial intelligence research, 22:457-479.
Jenny Rose Finkel, Trond Grenager, and Christopher Manning. 2005. Incorporating non-local information into information extraction systems by gibbs sampling. In Proceedings of the 43rd annual meeting on association for computational linguistics, pages 363-370. Association for Computational Linguistics.
Annemarie Friedrich, Marina Valeeva, and Alexis Palmer. 2014. LQVSumm: A corpus of linguistic quality violations in multi-document summarization. In Proceedings of the Ninth International Conference on Language Resources and Evaluation (LREC’14), pages 1591-1599, Reykjavik, Iceland. European Language Resources Association (ELRA).
Alexios Gidiotis and Grigorios Tsoumakas. 2019. Structured summarization of academic publications. In Joint European Conference on Machine Learning and Knowledge Discovery in Databases, pages 636645. Springer.
Jiatao Gu, Zhengdong Lu, Hang Li, and Victor OK Li. 2016. Incorporating copying mechanism in sequence-to-sequence learning. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers).
Anette Hulth. 2003. Improved automatic keyword extraction given more linguistic knowledge. In Proceedings of the 2003 conference on Empirical methods in natural language processing, pages 216-223. Association for Computational Linguistics.
Chris Kedzie, Kathleen McKeown, and Hal Daumé III. 2018. Content selection in deep learning models of summarization. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, pages 1818-1828.
Chin-Yew Lin. 2004. ROUGE: A package for automatic evaluation of summaries. In Text Summarization Branches Out, pages 74-81, Barcelona, Spain. Association for Computational Linguistics.
Mark F. Medress, Franklin S Cooper, Jim W. Forgie, CC Green, Dennis H. Klatt, Michael H. O’Malley, Edward P Neuburg, Allen Newell, DR Reddy, B Ritea, et al. 1977. Speech understanding systems: Report of a steering committee. Artificial Intelligence, 9(3):307-316.
Haitao Mi, Baskaran Sankaran, Zhiguo Wang, and Abe Ittycheriah. 2016. Coverage embedding models for neural machine translation. In Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, pages 955-960.
Rada Mihalcea and Paul Tarau. 2004. Textrank: Bringing order into text. In Proceedings of the 2004 conference on empirical methods in natural language processing, pages 404-411.
Ramesh Nallapati, Bowen Zhou, Cicero dos Santos, Çağlar Gulçehre, and Bing Xiang. 2016. Abstractive text summarization using sequence-to-sequence rnns and beyond. In Proceedings of The 20th SIGNLL Conference on Computational Natural Language Learning, pages 280-290.
Shashi Narayan, Shay B Cohen, and Mirella Lapata. 2018. Don’t give me the details, just the summary! topic-aware convolutional neural networks for extreme summarization. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, pages 1797-1807.
Karl Pearson. 1895. Vii. note on regression and inheritance in the case of two parents. proceedings of the royal society of London, 58(347-352):240-242.
Alexander M Rush, Sumit Chopra, and Jason Weston. 2015. A neural attention model for abstractive sentence summarization. In Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing, pages 379-389.
Abigail See, Peter J Liu, and Christopher D Manning. 2017. Get to the point: Summarization with pointergenerator networks. In Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), pages 10731083.
Burr Settles. 2005. Abner: an open source tool for automatically tagging genes, proteins and other entity names in text. Bioinformatics, 21(14):3191-3192.
Eva Sharma, Chen Li, and Lu Wang. 2019. Bigpatent: A large-scale dataset for abstractive and coherent summarization. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, pages 2204-2213.
Michael Völske, Martin Potthast, Shahbaz Syed, and Benno Stein. 2017. Tl; dr: Mining reddit to learn automatic summarization. In Proceedings of the Workshop on New Frontiers in Summarization, pages 5963.
A Summarization Example on CNN/DailyMail Dataset
We see factual redundancy and repetitiveness in the generated summaries with pointer-generation which is removed by applying coverage. In the example below the Factual Redundancy is shown with the bold text:
Reference Summary maricopa county sheriff 's office in arizona says robert bates never trained with them. " he met every requirement, and all he did was give of himself, "his attorney says. tulsa world newspaper: three supervisors who refused to sign forged records on robert bates were reassigned.
Summary from seq2seq some supervisors at the tulsa county sheriff’s office were told to forge reserve deputy robert bates ’ training records. some supervisors at the tulsa county sheriff’s office were told to forge reserve deputy robert bates’ training records, and three who refused were reassigned to less desirable duties. some supervisors at the tulsa county sheriff’s office were told to forge reserve deputy robert bates ’ training records.
Summary from seq2seq with coverage some supervisors at the tulsa county sheriff 's office were told to forge reserve deputy robert bates ’ training records . the volunteer deputy 's records had been falsified emerged almost immediately " from multiple sources after bates killed eric harris on april 2 . bates claims he meant to use his taser but accidentally fired his handgun at harris instead.
B Example of Summarization on SUMPubMED
Here we provide representative examples of actual summaries. Repetitiveness, i.e., factual redundancy is shown with the bold text.
B. 1 Abstractive Summarization on
SUMPubMED
We see factual redundancy and repetitiveness in the generated summaries with pointer-generation which is removed by applying coverage. We also observe that repetitiveness is removed by using the coverage mechanism.
reference: the origin of these genes has been attributed to horizontal gene transfer from bacteria, although there still is a lot of uncertainty about the origin and structure of the ancestral gbf < dig > ppn endoglucanase. our data confirm a close relationship between pratylenchus spp. furthermore, based on gene structure data, we inferred a model for the evolution of the gbf < dig > endoglucanase gene structure in plantparasitic nematodes. our evolutionary model for the gene structure in ppn gbf < dig > endoglucanases implies the occurrence of an early duplication event, and more recent gene duplications at genus or species level. the latter one is the first gene isolated from a ppn of a different superfamily -LRB- sphaerularioidea -RRB-; all previously known nematode endoglucanases belong to the superfamily tylenchoidea -LRB- order rhabditida -RRB-. no statistical incongruence between the phylogenetic trees deduced from the catalytic domain and the cbm<dig> was found, which could suggest that both domains have evolved together. and the root knot nematodes, while some radopholus similis endoglucanases are more similar to cyst nematode genes. two new endoglucanases from the migratory nematodes pratylenchus coffeae and ditylenchus africanus were included in this study. phylogenetic analyses were conducted with the ppn gbf < dig > endoglucanases and homologous endoglucanases from bacterial and other eukaryotic lineages such as beetles, fungi and plants. previous studies on the evolution of these genes have focused primarily on data of sedentary nematodes, while in this study, extra data from migratory nematodes were included. it is not clear whether this ancestral endoglucanase consisted of the whole gene cassette, containing a catalytic domain and a carbohydratebinding module -LRB- cbm, type < dig > in ppn and bacteria -RRB- or only of the catalytic domain while the cbm<dig> was retrieved by domain shuffling later in evolution. the gbf < dig > catalytic domain and the cbm < dig > rather than that it evolved by domain shuffling. endo < dig > betaglucanases or cellulases from the glycosyl hydrolase family < dig >-LRB- gbf5 -RRB- have been found in numerous bacteria and fungi, and recently also in higher eukaryotes, particularly in plantparasitic nematodes -LRB- ppn -RRB-.
seq2seq: although the hox cell line with peculiar properties of horizontal gene transfer -LRB- hgt -RRB- has been proposed to delineate the function of ghf < dig > endoglucanases. the gene structure of the ppn ghf < dig > endoglucanases is a matter of debate, in this study no support for an early exon/domain shuffling event after the independent gain of a catalytic domain. the exon/intron gene structure of the ppn ghf < dig > endoglucanases was compared with the multiple protein sequence alignment. the gene structure of the ppn ghf < dig > endoglucanases was compared with the multiple protein sequence alignment. the gene structure of the ppn ghf < dig > endoglucanases was compared with the multiple protein sequence alignment. the gene structure of the ppn ghf < dig > endoglucanases was compared with the multiple protein sequence alignment. the evolution of the gene structure of the ppn ghf < dig > gene families: the number of members from an ancient or early eukaryotic ancestral gene is associated with the expansion of members from an ancient or early eukaryotic ancestral gene. in this study, we.
seq2seq coverage: the expansion of horizontal gene transfer-LRB- hgt -RRB- events in horizontal gene transfer -LRB- hgt -RRB- has been proposed to explain the origin of ghf < dig > endoglucanases in the nematode kingdom. while the ppn ghf < dig > endoglucanases has a close relationship to the root knot nematodes. in order to have a broader overview of the endoglucanase evolution in the infraorder tylenchomorpha, the gene structure of six additional genes was incorporated in our study. the ppn ghf < dig > gene family is associated with the expansion of the ppn ghf < dig > gene family bordered by intron < dig > and intron < dig > although 1−< dig > symmetrical domains are suggested to be frequently associated with domain shuffling events in the evolution of paralogous gene families: the evolution of the ppn indicate a history of recent duplication events for which little information is available. our model implies that the divergence of the gene structure of the ppn ghf < dig > gene family is notably dynamic, and this evolution involves more intron gains than losses in the order rhabditida-LRB- infraorder tylenchomorpha-RRB-, which is part of one of the three evolutionary independent plantparasitic nematode clades. our results demonstrate that the conserved gene structure of the ppn ghf < dig > endoglucanases and the observation of some sequence conservation in the evolution of the plantparasitic bacteria and nematodes. our results suggest that the evolution of the ghf < dig > gene family is a major consequence of the evolution of.
B. 2 Extractive Summarization on SUMPUBMED
TextRank produces a purely extractive summary. But we see that it is able to identify the relevant sentences. The content overlap between the reference and generated extractive summary is adequate.
reference : to find out the different ovarian activity and follicle recruitment with mirnamediated posttranscriptional regulation, the small rnas expressed pattern in the ovarian tissues of multiple and uniparous anhui white goats during follicular phase was analyzed using solexa sequencing data. < dig > mirnas coexpressed, < dig > and < dig > mirnas specifically expressed in the ovaries of multiple and uniparous goats during follicular phase were identified. in the present study, the different expression of mirnas in the ovaries of multiple and uniparous goats during follicular phase were characterized and investigated using deep sequencing technology. rt-pcr was applied to detect the expression level of < dig > randomly selected mirnas in multiple and uniparous hircine ovaries, and the results were consistent with the solexa sequencing data. micrornas play critical roles in almost all ovarian biological processes, including folliculogenesis, follicle development, follicle atresia, luteal development and regression. the result will help to further understand the role of mirnas in kidding rate regulation and also may help to identify mirnas which could be potentially used to increase hircine ovulation rate and kidding rate in the future. the < dig > most highly expressed mirnas in the multiple library were also the highest expressed in the uniparous library, and there were no significantly different between each other. the highest specific expressed mirna in the multiple library was mir29c, and the one in the uniparous library was mir < dig >< dig > novel mirnas were predicted in total. superior kidding rate is an important economic trait in production of meat goat, and ovulation rate is the precondition of kidding rate. go annotation and kegg pathway analyses were implemented on target genes of all mirna in two libraries.
extracted : in order to identify differentially expressed mirna during follicular phase in the ovaries of multiple and uniparous anhui white goats, two small rna libraries were constructed by solexa sequencing, for all mirnas target genes of multiple and uniparous goats in the ovaries during follicular phase, there were <dig> and <dig> target genes mapped to the go terms of cellular component. the expression levels of <dig> randomly selected mirnas were verified in the ovaries of multiple and uniparous goats during follicular phase using rt-pcr. in this study, we sequenced the small rnas in the ovarian tissues of multiple and uniparous anhui white goats during follicular phase by illumina solexa technology, then analyzed the differentially expressed mirnas, predicted novel mirnas, and made go enrichment and kegg pathway analysis of target genes in two mirna libraries. in ovaries between multiple and uniparous goats of follicular phase, < dig > novel mirnas were predicted in total, which is distinctly more than the amount predicted in our previous study implemented by our team workers, zhang et al. the highest specific expressed mirna in multiple library was mir29c, and the one in uniparous library was mir < dig > as aligning the clean reads to the mirna precursor/mature mirnas of all animals in the mirbase < dig > database, and obtained mirna with no specified species. rt-pcr was carried out to analyze the expression of < dig > randomly selected mirnas in multiple and uniparous hircine ovaries during follicular phase, and the results were consistent with the solexa sequencing data.
B. 3 Attention Visualization for SUMPUBMED
We can visualize the attention projection for seq2seq models by highlighting the respective words in yellow on the source document while producing a word. Figures 2 and 3 show the words in green with high generation probability, i.e, pgen >0.5 (not copied), non marked words have pgen <0.5 (mostly copied).
Observations While producing a word in the output, we can visualize the respective words in the source document on which the network is focussing. The darker the green highlight over a word in the summary, the higher is the pgen prob- ability. E.g., there is a chance that pgen is high whenever a new sentence is started after a period (.). The model generally focuses on two or three words at a time. There is a high chance that the summary starts with a noun phrase or a noun. For example, we can see in Figure 2 that the summary starts with name (noun) ‘kevin pietersen’.
Article
it 's the picture some england cricket fans have been waiting to see and others have been equally dreading : kevin pietersen back at survey. The 3d-per-old returned to nets on monday for the first time since. re-signing. for the county last month he arrived early for the year. leaming a picture of the picture was the same. a s evel before leam-extra such as gerath bets and jade. Berdson. followed his in. kevin pietersen is pictured leaving the oval for the first time since resigning for survey last month. pietersentreturned to nets at survey on monday and left the oval after training finished just before jom. pietersences pictured driving away from the oval in his expensive. .teisa. sports car. pietersencesaged a any seile as he drove away after training on monday afternoon. pietersent was later pictured leaving the ground just before jom and is expected to. .srep-up. his county rehabilitation with a three-day were-up against seferal. .mics. on april 12 . ultimately. pietersent is hoping to impress enough for survey to earn a. re-call. to the england side possibly for this summer 's ashes remain. having been sacked by the national sum in 2014. england left for the west indaes for their upcoming test series on thursday. with touch peter mores leaving 65 in no doubt that he still has a lot to prove despite incoming england and wales cricket board chairman color graces appearing to extended on olive branch to the exiled batseun. asked at getwick about pietersen 's situation. moores said: from my point of view, kevin is n’t on the radar.
Reference summary
kevin pietersen took part in a net session at the oval on monday. he is expected to play in three-day game against seferal. .mics. on april 12 . pietersen has returned to county game to boost chances of england recall.
Generated summary (highlighted = high generation probability)
kevin pietersen returned to nets on monday for the first time since resigning for survey last month. She returned to nets at survey on monday for left the oval after training on monday. a pietersent is hoping to impress enough for survey to earn a re-call to the england side.
Figure 2: Attention Probability for decoding on DUC 2001 dataset example, showing the summary is more inclined to an extractive nature. Attention corresponding to the word ‘pietersen’ present in the generated summary is shown.
Article
In line with these results, net studies using transient reduction of tinnitus by lidocaine also revealed significantly increased. roff. in. temporoparietal cortices activity during tinnitus perception ’ regarding cortical excitability memory. significantly reduced occipitalisth, Yellinterior and Temporal Tinnitus activity during cianthation. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
temporal and temporal tinnitus activity at low frequencies in the temporal fimbria. single anterior of the net mations at high frequencies are results of a. nnery testisn. but
References (21)
- Wei-Fan Chen, Shahbaz Syed, Benno Stein, Matthias Hagen, and Martin Potthast. 2020. Abstractive snip- pet generation. In Proceedings of The Web Confer- ence 2020, pages 1309-1319.
- Arman Cohan, Franck Dernoncourt, Doo Soon Kim, Trung Bui, Seokhwan Kim, Walter Chang, and Nazli Goharian. 2018. A discourse-aware attention model for abstractive summarization of long documents. In Proceedings of the 2018 Conference of the North American Chapter of the Association for Computa- tional Linguistics: Human Language Technologies, Volume 2 (Short Papers), volume 2, pages 615-621.
- Arman Cohan and Nazli Goharian. 2016. Revisiting summarization evaluation for scientific articles. In Proceedings of the Tenth International Conference on Language Resources and Evaluation (LREC'16), pages 806-813.
- Shibhansh Dohare, Harish Karnick, and Vivek Gupta. 2017. Text summarization using abstract meaning representation. arXiv preprint arXiv:1706.01678.
- Günes Erkan and Dragomir R Radev. 2004. Lexrank: Graph-based lexical centrality as salience in text summarization. Journal of artificial intelligence re- search, 22:457-479.
- Jenny Rose Finkel, Trond Grenager, and Christopher Manning. 2005. Incorporating non-local informa- tion into information extraction systems by gibbs sampling. In Proceedings of the 43rd annual meet- ing on association for computational linguistics, pages 363-370. Association for Computational Lin- guistics.
- Annemarie Friedrich, Marina Valeeva, and Alexis Palmer. 2014. LQVSumm: A corpus of linguis- tic quality violations in multi-document summariza- tion. In Proceedings of the Ninth International Conference on Language Resources and Evalua- tion (LREC'14), pages 1591-1599, Reykjavik, Ice- land. European Language Resources Association (ELRA).
- Alexios Gidiotis and Grigorios Tsoumakas. 2019. Structured summarization of academic publications. In Joint European Conference on Machine Learning and Knowledge Discovery in Databases, pages 636- 645. Springer.
- Jiatao Gu, Zhengdong Lu, Hang Li, and Victor OK Li. 2016. Incorporating copying mechanism in sequence-to-sequence learning. In Proceedings of the 54th Annual Meeting of the Association for Com- putational Linguistics (Volume 1: Long Papers).
- Anette Hulth. 2003. Improved automatic keyword ex- traction given more linguistic knowledge. In Pro- ceedings of the 2003 conference on Empirical meth- ods in natural language processing, pages 216-223. Association for Computational Linguistics.
- Chris Kedzie, Kathleen McKeown, and Hal Daumé III. 2018. Content selection in deep learning models of summarization. In Proceedings of the 2018 Con- ference on Empirical Methods in Natural Language Processing, pages 1818-1828.
- Chin-Yew Lin. 2004. ROUGE: A package for auto- matic evaluation of summaries. In Text Summariza- tion Branches Out, pages 74-81, Barcelona, Spain. Association for Computational Linguistics.
- Mark F. Medress, Franklin S Cooper, Jim W. Forgie, CC Green, Dennis H. Klatt, Michael H. O'Malley, Edward P Neuburg, Allen Newell, DR Reddy, B Ritea, et al. 1977. Speech understanding systems: Report of a steering committee. Artificial Intelli- gence, 9(3):307-316.
- Haitao Mi, Baskaran Sankaran, Zhiguo Wang, and Abe Ittycheriah. 2016. Coverage embedding models for neural machine translation. In Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, pages 955-960.
- Rada Mihalcea and Paul Tarau. 2004. Textrank: Bring- ing order into text. In Proceedings of the 2004 con- ference on empirical methods in natural language processing, pages 404-411.
- Ramesh Nallapati, Bowen Zhou, Cicero dos Santos, C ¸aglar Gulc ¸ehre, and Bing Xiang. 2016. Abstrac- tive text summarization using sequence-to-sequence rnns and beyond. In Proceedings of The 20th SIGNLL Conference on Computational Natural Lan- guage Learning, pages 280-290.
- Shashi Narayan, Shay B Cohen, and Mirella Lapata. 2018. Don't give me the details, just the summary! topic-aware convolutional neural networks for ex- treme summarization. In Proceedings of the 2018 Conference on Empirical Methods in Natural Lan- guage Processing, pages 1797-1807.
- Karl Pearson. 1895. Vii. note on regression and inheri- tance in the case of two parents. proceedings of the royal society of London, 58(347-352):240-242.
- Alexander M Rush, Sumit Chopra, and Jason Weston. 2015. A neural attention model for abstractive sen- tence summarization. In Proceedings of the 2015 Conference on Empirical Methods in Natural Lan- guage Processing, pages 379-389.
- Abigail See, Peter J Liu, and Christopher D Manning. 2017. Get to the point: Summarization with pointer- generator networks. In Proceedings of the 55th An- nual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), pages 1073- 1083.
- Burr Settles. 2005. Abner: an open source tool for au- tomatically tagging genes, proteins and other entity names in text. Bioinformatics, 21(14):3191-3192.
 Harish Karnick
Harish Karnick